

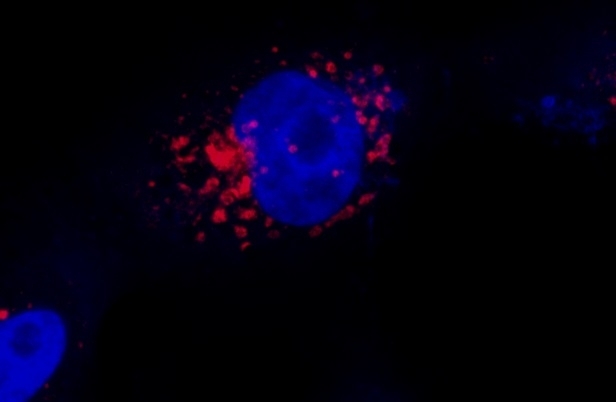
Protocol developed by Brazilian researchers shows SARS-CoV-2 replicating near cell nucleus. Methodology helps scientists understand coronavirus’s action mechanism and could also be used to study other viruses (image: Luana Nunes Santos / UNICAMP)
Published on 03/18/2021
By André Julião | Agência FAPESP – Researchers at the University of Campinas (UNICAMP) in the state of São Paulo, Brazil, have developed a method of displaying the genetic material of the novel coronavirus inside cells. Based on a technique known as fluorescent in situ hybridization (FISH), it exhibits 3D images of the virus as well as other cell components.
“Laboratories typically use techniques like qPCR that measure viral load in cultured cells or infected tissue, but these techniques can’t prove the virus is in the cells or specify which part of the cell it penetrates. This type of precision is important to an understanding of the disease,” said Henrique Marques-Souza, a professor in the university’s Biology Institute (IB-UNICAMP) and leader of the team that developed the method.
Marques-Souza is supported by FAPESP and is a member of UNICAMP’s COVID-19 task force, which brings together research efforts, test materials, and resources to understand and combat the disease.
The protocol, which was developed by postdoctoral fellow Luana Nunes Santos, can be used to deepen the research on the novel coronavirus already in progress at their lab and facilitate collaboration with other researchers at UNICAMP and elsewhere (read more at: agencia.fapesp.br/32998/).
“Being able to visualize the virus inside the cell is very valuable to help us understand how infection works,” Marques-Souza said. “It can also be visualized using a transmission electron microscope [TEM] or immunocytochemistry [ICC]. However, TEM requires a special microscope and takes between a week and 10 days to complete. ICC requires antibodies that bind to the virus. It’s relatively simple, but the assay materials are expensive and take a long time to arrive owing to high world demand during the pandemic.”
In FISH, the researchers synthesize a probe – a short DNA sequence with a fluorescent molecule attached to it. When it comes into contact with an infected cell, it binds (hybridizes) to the viral RNA and the fluorescent molecule makes it visible under a fluorescence microscope.
From the logistical standpoint, the assays can be performed faster by the UNICAMP team because they do not depend on imports of commercial FISH kits or of antibodies for immunocytochemistry, and this also represents a financial benefit. Another advantage is that the virus can be detected sooner since immunocytochemistry depends only on viral RNA replication to a detectable level.
The study was conducted in partnership with the Laboratory for Research on Emerging Viruses (LEVE), led by José Luiz Proença Módena, and the National Institute of Science and Technology (INCT) in Photonics Applied to Cellular Biology (INFABiC), one of the INCTs supported by FAPESP in the state of São Paulo together with Brazil’s National Council for Scientific and Technological Development (CNPq).
INFABiC’s principal investigator is Hernandes Carvalho. Módena and Carvalho are professors at IB-UNICAMP. The project is also supported by UNICAMP’s Pro-Rector for Research via the Faepex fund.
Viral action
The 3D images produced so far show that the virus replicates near the cell nucleus and probably penetrates specific organelles such as endosomes. The researchers are using the technique to answer a range of questions about the viral infection mechanism, and the first results are soon to be submitted for publication.
The research also offers a way forward for further research on other viruses, not least in order to see if there are parallels with SARS-CoV-2. “Everything we discovered about the dynamics of the virus in the cell can be adapted for comparison with influenza and other more commonplace viruses. This may help explain why SARS-CoV-2 is so aggressive,” Marques-Souza said.
The group also expect to develop a novel test to detect the virus in future, although this is not their focus right now. The protocol has been tested on Vero cells, derived from the kidney of an African monkey and the most widely used model in coronavirus studies. The assays were also successful in lung cells and other human cells, evidencing the versatility of the technique.
Source: https://agencia.fapesp.br/34248