

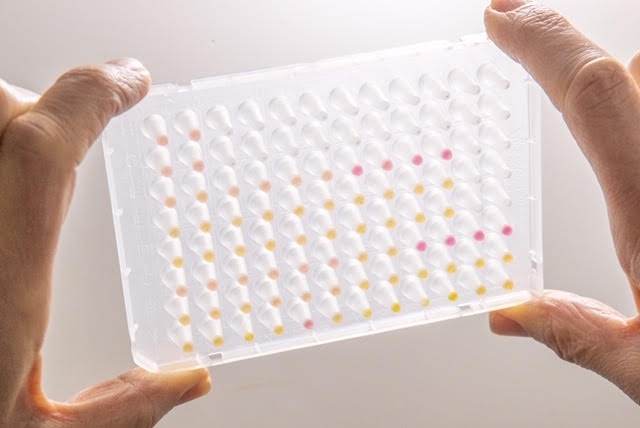
Designed by the Human Genome and Stem Cell Research Center, the novel test may cost a quarter of those based on RT-PCR, considered the gold standard for diagnosis of the disease (multi-well plate with test results: yellow dots are positives, pink dots negative / image: HUG-CELL)
Published on 03/18/2021
By Elton Alisson | Agência FAPESP – In Brazil, researchers at the Human Genome and Stem Cell Research Center (HUG-CELL), hosted by the University of São Paulo (USP), are about to complete the development of a saliva-based COVID-19 diagnostic test.
The test may cost a quarter of those based on RT-PCR (reverse transcription polymerase chain reaction), considered the gold standard for diagnosis of the disease and currently performed by laboratories in Brazil for the equivalent of USD65-75.
HUG-CELL is linked to USP’s Bioscience Institute (IB) and is one of the Research, Innovation and Dissemination Centers (RIDCs) funded by FAPESP.
“Like RT-PCR, the alternative test will be used to detect the virus during infection,” said Maria Rita Passos-Bueno, a member of the HUG-CELL research team and principal investigator for the project.
The test is similar in concept to those already developed in Brazil and elsewhere with the aim of increasing the availability and speed of molecular testing, as well as lowering its cost, by simplifying the process.
In the United States, for example, the Food and Drug Administration (FDA) has so far given Emergency Use Authorization to five saliva-based COVID-19 diagnostic tests. The latest is SalivaDirect, developed by researchers at Yale University.
In Brazil, genomics company Mendelics is offering its own saliva-based test, and researchers at the Federal University of Goiás are developing a diagnostic kit along similar lines.
Simpler method
The new tests are based on RT-LAMP (reverse transcription loop-mediated isothermal amplification), a molecular technique widely used to diagnose infectious diseases caused by RNA viruses such as dengue, chikungunya, hepatitis A and zika.
It is similar in some ways to RT-PCR, which requires sampling by nasopharyngeal swab. Both use chemical reagents to perform reverse transcription (RT), in which the viral RNA is converted into DNA, followed by amplification to replicate specific regions of DNA millions of times in order to identify the pathogen.
However, RT-LAMP does not require the extraction of RNA from the virus to be detected. In the case of RT-PCR this is necessary and is done by reagents, which Brazil has to import, so they are expensive and often hard to find on the market, depending on demand. Another advantage of RT-LAMP is that it does not use complex laboratory equipment like the real-time thermocycler used in RT-PCR to detect and amplify RNA by exposing the sample to different temperatures.
“In RT-LAMP, amplification is done in a single step without the need to extract RNA. In addition, the process can be performed at a fixed temperature, say 65 °C, using simple equipment such as a water bath. This also simplifies the process and shortens the time taken to get the results,” Passos-Bueno said.
RNA extraction is time-consuming and complex. Elimination of this step in RT-LAMP saves time and money. “In RT-PCR the RNA extraction step can take between one and five hours depending on the system used,” she explained. “Skipping this stage cuts the cost of the RT-LAMP test by 30%.”
The group are able to eliminate RNA extraction in RT-LAMP by heating the viral capsid until it bursts and adding a solution developed by them to stabilize the virus for conversion of its RNA into DNA and amplification to facilitate detection in saliva. The entire process takes place at a constant temperature (hence the term isothermal in the name of the technique).
The enzymes required for this process are also imported and represent the largest proportion of its cost, but substitutes have been developed by a group of researchers in USP’s Chemistry Institute (IQ) led by Professor Shaker Chuck Farah. “We’ve assured indigenous production of the enzymes, the main inputs for the test, and this will further lower its cost,” Passos-Bueno said.
Standardization
The researchers now plan to standardize the test by using chemical solutions that keep the viral RNA stable for a long period so that it is not affected by the enzymes present in saliva.
“Saliva contains several substances that can inhibit the action of the enzymes, break down the viral material and interfere with the amplification reaction, so we’re developing buffers to enable users to standardize the conditions under which saliva samples and viral RNA are stored, and to ensure the test is performed with as low as possible a risk of false negatives,” Passos-Bueno said.
The test has exhibited 100% specificity for detection of the novel coronavirus, matching conventional tests. The researchers want to enhance its sensitivity so that it detects the virus in a very small number of copies derived from saliva.
“Even now its sensitivity is good: it can detect up to ten copies per reaction in saliva samples,” Passos-Bueno said.
The patient can be responsible for collection of the saliva sample in a test tube. The procedure is painless and non-invasive, avoiding the involvement of suitably appareled health workers trained to sample nasopharyngeal secretion, as in RT-PCR testing. Hence it reduces the risk of contagion and avoids swabs altogether.
The researchers believe the result will be ready within 30-40 minutes. “The result is visible to the naked eye. The tube containing the sample changes color if the virus is present. A negative result is pink and a positive is yellow,” Passos-Bueno said.
One of the project’s goals is offering the test in places lacking in infrastructure for collection and analysis by means of inclusion of university-based reference laboratories to expand Brazil’s testing capacity.
Source: https://agencia.fapesp.br/34135