

Researchers at the University of São Paulo found that the SARS-CoV-2 variant’s spike protein interacted more strongly with the ACE2 receptor used by the virus to invade and replicate in human cells (image: Jadson Santos / USP)
Published on 03/15/2021
By Elton Alisson | Agência FAPESP – Researchers at the University of São Paulo’s Ribeirão Preto Medical School (FMRP-USP) and Dental School (FORP-USP) in Brazil have identified one of the factors that increase the transmissibility of the novel SARS-CoV-2 variant known as B.1.1.7. This variant was first detected in the United Kingdom.
Two cases of COVID-19 caused by the variant have so far been confirmed in Brazil by Adolf Lutz Institute, São Paulo State’s central public health laboratory.
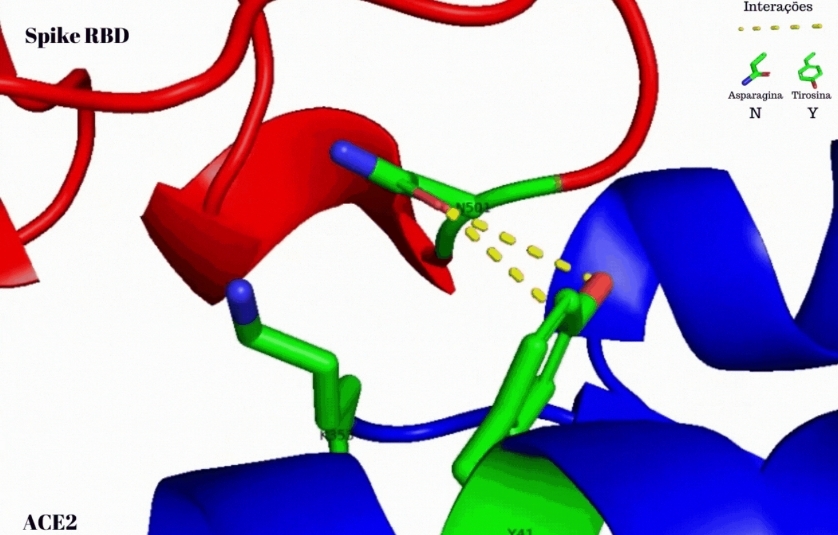
The researchers used bioinformatics tools to prove their hypothesis that the novel variant’s spike protein, which mediates viral entry into host cells, establishes stronger molecular interaction with the ACE2 receptor on the cell surface. The increase in interaction force is due to a mutation (N501Y) that increases the virus’s binding affinity to ACE2.
The study was supported by FAPESP and is reported in a paper posted to the biology repository bioRxiv. The paper is a preprint and has not yet been certified by peer review.
“We found that interaction between the spike protein of the novel variant and the N501Y mutation is much stronger than that of the first lineage of the virus isolated in Wuhan, China,” Geraldo Aleixo Passos, last author of the paper and principal investigator for the study, told Agência FAPESP. Passos is a professor at FMRP-USP and FORP-USP. His co-author, Jadson Santos, who performed the bioinformatics analysis, is a PhD candidate at FMRP-USP whose research is supervised by Passos.
When B.1.1.7 was identified in the UK, the researchers hypothesized that the mutation in the spike protein, written N501Y to denote the substitution of tyrosine (Y) for asparagine (N) at site 501 in the amino acid sequence, could be one of the factors responsible for the novel variant’s greatly increased transmissibility.
N501 had already been identified as a crucial amino acid residue in the spike protein’s binding affinity to ACE2 and hence as being involved in the infectivity of the virus. Previous research had also shown that the mutation N501Y found in B.1.1.7 covers one of the six key contact amino acid residues in the spike protein.
“There are other mutations in the genome of this lineage which we didn’t analyze. We focused on N501Y because of its involvement in the interaction between the spike and ACE2,” Passos explained.
To test the hypothesis that the high infectivity of B.1.1.7 was due to changes in the force of spike-ACE2 interaction, the researchers used structures from the spikes of the original Wuhan reference strain and B.1.1.7 deposited in the Protein Data Bank. They used an open-source molecular visualization system called PyMOL to view the interaction between the N501 amino acid residue of the SARS-CoV-2 (Wuhan) spike and the Y41 residue of the human ACE2 receptor protein, and to simulate and analyze the interactions resulting from the N501Y mutation in B.1.1.7.
“The software system enabled us to view images of these molecular structures at a resolution of 3.5 angstrom, representing a magnification even greater than you can get from an ultramicroscope,” Passos said.
The researchers used another open-source bioinformatics tool called PDBePISA to compare the interaction between the wild (original) and mutant strains’ spike proteins and ACE2.
The results of the analyses showed that the N501Y mutation in the novel variant’s spike interacted more with ACE2 than the wild strain of the virus. The interactions were predominantly non-covalent (weaker).
“The sum of the various weak interactions between the novel variant’s mutant spike protein and the human ACE2 receptor results in stronger molecular interaction, so that the virus can more easily enter and replicate in cells,” Passos said.
The N501Y mutation also caused a change in the spacing between the amino acid residues in the spike protein, enabling them to establish more interactions with ACE2.
“Together these changes confirmed the hypothesis that the B.1.1.7 strain’s spike protein interacts more strongly with the ACE2 receptor,” Passos said.
The results of the in silico study using computer simulations, he added, can now serve as a basis for other in vitro experiments designed to investigate the infectivity of the new variant using human cells cultured in the laboratory.
Surprising evolution
According to the researchers, the rapid spread of SARS-CoV-2 among humans is driving its molecular evolution. To date the virus has mutated at a rate of up to two nucleotides per month, and at least 20 nucleotide alterations have been found in the genomes of recent isolates compared with the wild lineage isolated in January 2020. Most of the mutations are located in the spike protein.
Lineage B.1.1.7, detected in early September, described in December 2020 by the COVID-19 Genomics UK (COG-UK) Consortium, and since reported in 17 other countries including Brazil, is one of several examples of this rapid evolution, but scientists were surprised to find that it has 17 mutations, eight of which are located in the gene that encodes the spike protein.
“The novel lineage has many more mutations than other lineages of the coronavirus,” Passos said.
Because the variant has been described so recently, it is too soon to say whether its phenotype makes it more or less virulent (disease-causing), he explained.
The following video shows how the spike protein of the new viral strain interacts with the ACE2 receptor:
The article “The high infectivity of SARS-CoV-2 B.1.1.7 is associated with increased interaction force between Spike-ACE2 caused by the viral N501Y mutation” (doi: 10.1101/2020.12.29.424708) by Jadson C. Santos and Geraldo A. Passos can be read at: www.biorxiv.org/content/10.1101/2020.12.29.424708v1.full.
Source: https://agencia.fapesp.br/35021